Science
Deep Learning Transforms 3D Imaging of Fruit Tissue Microstructure
A recent study published in the journal Plant Phenomics on July 5, 2025, reveals a groundbreaking advancement in the imaging of fruit tissues. Researchers from KU Leuven, led by Pieter Verboven, successfully developed a deep learning model that significantly enhances the accuracy of 3D segmentation of apple and pear tissues, surpassing previous 2D methods.
Traditional microscopy techniques have long posed challenges for studying the 3D structure of plant tissues, essential for understanding various metabolic processes. These methods require extensive sample preparation and typically provide limited fields of view. Although X-ray micro-CT has facilitated non-destructive 3D imaging, quantifying tissue morphology remains complex due to overlapping features and poor image contrast. Existing segmentation methods often struggle to accurately identify parenchyma cells, vascular tissues, and stone cell clusters.
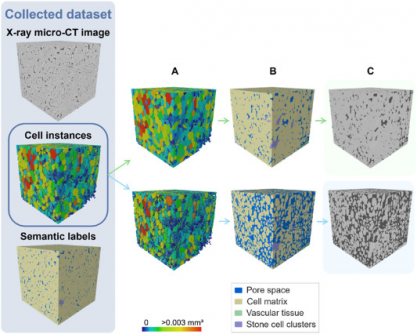
Recognizing these limitations, Verboven’s team sought to create an automated framework for labeling and quantifying plant tissue architecture. The research introduces a 3D panoptic segmentation framework that integrates the 3D extension of Cellpose and a 3D Residual U-Net for detailed analysis of fruit tissue microstructure from X-ray micro-CT images.
The model employs both instance segmentation and semantic segmentation techniques. It predicts intermediate gradient fields in three dimensions to distinguish individual parenchyma cells while classifying voxels into various categories, including cell matrix, pore space, vasculature, and stone cell clusters. The training involved datasets of apples and pears, augmented with synthetic data to improve model robustness.
Performance metrics demonstrated remarkable results. The model achieved an Aggregated Jaccard Index (AJI) of 0.889 for apples and 0.773 for pears, outperforming the previous 2D model, which had AJIs of 0.861 and 0.732, respectively. Its capabilities in accurately segmenting pore spaces and cell matrices were particularly notable, while it also identified vasculature with a Dice Similarity Coefficient (DSC) of 0.506 for apples and 0.789 for pears. The model demonstrated robust performance in segmenting stone cell clusters as well, achieving a precision of 0.798 and a recall of 0.836.
Visual validation further confirmed the model’s efficacy, accurately detecting vascular bundles in varieties such as ‘Kizuri’ and ‘Braeburn’ apples. The segmentation of stone cell clusters in ‘Celina’ and ‘Fred’ pears also exhibited realistic results, with DSC values reaching up to 0.90. Despite these successes, the researchers noted that additional data augmentation did not significantly enhance performance, likely due to dataset imbalance and domain shifts.
Morphometric analysis provided further validation of the model’s accuracy, revealing vasculature widths ranging from 70–780 μm and variability in stone cell cluster dimensions and sphericity, which measured between 0.68–0.74. This 3D deep learning approach represents a substantial advancement in the field, offering a complete, automated, and contrast-free method for quantifying plant tissue microstructure.
The implications of this research extend beyond the immediate findings. The model serves as a powerful, non-destructive tool for plant scientists, enabling them to study how microscopic structures influence water, gas, and nutrient transport. This technology could significantly streamline “human-in-the-loop” analysis, minimizing manual effort while enhancing accuracy in tissue characterization.
In the context of fruit research, understanding cellular arrangements is crucial for determining texture, storability, and susceptibility to physiological disorders such as browning or watercore. The framework also holds promise for broader applications in studying tissue development, ripening, and stress responses across various crops, making it a valuable resource in plant anatomy and food science research.
This innovative tool, compatible with standard X-ray micro-CT instruments, paves the way for integrating artificial intelligence into plant phenomics, thus contributing to advancements in agriculture, horticulture, and related scientific disciplines.
Funding for this research was provided by the Research Foundation – Flanders (FWO, grant number S003421N, SBO project FoodPhase) and KU Leuven (project C1 C14/22/076).
For further details, the study can be accessed via the DOI link: 10.1016/j.plaphe.2025.100087.
-

 Business1 week ago
Business1 week agoIconic Sand Dollar Social Club Listed for $3 Million in Folly Beach
-

 Politics1 week ago
Politics1 week agoAfghan Refugee Detained by ICE After Asylum Hearing in New York
-

 Health1 week ago
Health1 week agoPeptilogics Secures $78 Million to Combat Prosthetic Joint Infections
-

 Science1 week ago
Science1 week agoResearchers Achieve Fastest Genome Sequencing in Under Four Hours
-

 Lifestyle1 week ago
Lifestyle1 week agoJump for Good: San Clemente Pier Fundraiser Allows Legal Leaps
-

 Health1 week ago
Health1 week agoResearcher Uncovers Zika Virus Pathway to Placenta Using Nanotubes
-

 World1 week ago
World1 week agoUS Passport Ranks Drop Out of Top 10 for First Time Ever
-

 Entertainment1 week ago
Entertainment1 week agoJennifer Lopez Addresses A-Rod Split in Candid Interview
-

 World1 week ago
World1 week agoRegional Pilots’ Salaries Surge to Six Figures in 2025
-
 Science1 week ago
Science1 week agoMars Observed: Detailed Imaging Reveals Dust Avalanche Dynamics
-

 Top Stories6 days ago
Top Stories6 days agoChicago Symphony Orchestra Dazzles with Berlioz Under Mäkelä
-

 Business1 week ago
Business1 week agoMcEwen Inc. Secures Tartan Lake Gold Mine Through Acquisition