Science
New Technique Reveals Chromatin Fiber Structures in Cells
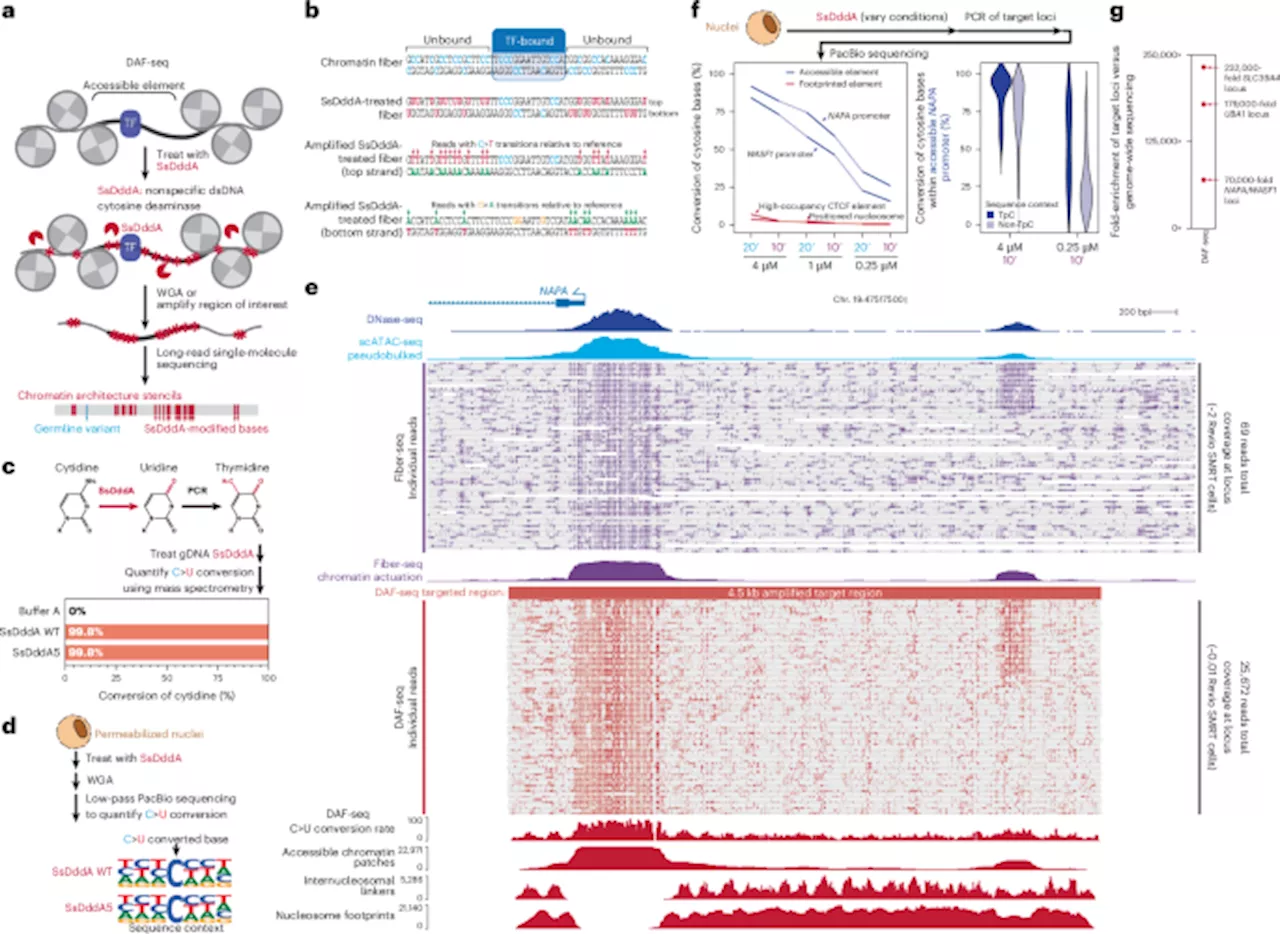
Researchers at the University of Washington School of Medicine have developed a groundbreaking technique known as Deaminase-Assisted single-molecule chromatin Fiber sequencing, or DAF-seq, which offers unprecedented insights into gene regulation at the single-cell level. This innovation allows scientists to map the intricate architectures of chromatin fibers, shedding light on how proteins bind to DNA within individual cells.
Gene regulation is complex, particularly in diploid organisms where two sets of chromosomes exist. The variability in protein occupancy along these chromatin fibers across different haplotypes and cells has been a significant challenge for researchers. DAF-seq addresses this gap by enabling single-molecule footprinting with near-nucleotide resolution, allowing the profiling of chromatin states and DNA sequences simultaneously.
With DAF-seq, researchers have discovered that cooperative protein occupancy occurs at individual regulatory elements, revealing the functional impact of somatic variants and rare chromatin epialleles. This technique generates comprehensive protein co-occupancy maps for approximately 99% of an individual cell’s mappable genome, providing a detailed view of chromatin plasticity within and between diploid cells.
Significant Findings from DAF-seq
The application of single-cell DAF-seq has uncovered extensive chromatin plasticity. Notably, researchers observed that chromatin actuation diverges by an astonishing 61% between haplotypes within a single cell, and by 63% between different cells. This variance suggests a dynamic regulatory landscape that can significantly influence gene expression.
Moreover, DAF-seq has shown that regulatory elements tend to co-actuate along the same chromatin fiber, following a distance-dependent pattern that mirrors the behavior of cohesin-mediated loops. This discovery emphasizes the intricate relationships between various regulatory elements, highlighting how spatial organization can impact gene regulation.
Overall, DAF-seq provides a powerful tool for characterizing protein occupancy across entire chromosomes with impressive precision—single-nucleotide, single-molecule, single-haplotype, and single-cell resolution. This advancement not only enhances our understanding of gene regulation but also opens new avenues for research into genetic diversity and disease mechanisms.
Researchers attribute the success of this technique to collaboration across multiple institutions, including the Edison Family Center for Genome Sciences and Systems Biology at Washington University in St. Louis. The study received funding from notable organizations such as the National Institutes of Health and the Chan Zuckerberg Initiative, demonstrating a strong commitment to advancing genomic research.
As the scientific community continues to explore the implications of DAF-seq, its potential applications in understanding diseases and personalized medicine could be transformative. The insights gained from this technique promise to deepen our understanding of the fundamental processes underlying gene regulation and chromatin architecture.
-

 Science1 month ago
Science1 month agoUniversity of Hawaiʻi Leads $25M AI Project to Monitor Natural Disasters
-
 Science2 months ago
Science2 months agoInterstellar Object 3I/ATLAS Emits Unique Metal Alloy, Says Scientist
-

 Science2 months ago
Science2 months agoResearchers Achieve Fastest Genome Sequencing in Under Four Hours
-

 Business2 months ago
Business2 months agoIconic Sand Dollar Social Club Listed for $3 Million in Folly Beach
-

 Politics2 months ago
Politics2 months agoAfghan Refugee Detained by ICE After Asylum Hearing in New York
-

 Business2 months ago
Business2 months agoMcEwen Inc. Secures Tartan Lake Gold Mine Through Acquisition
-

 Health2 months ago
Health2 months agoPeptilogics Secures $78 Million to Combat Prosthetic Joint Infections
-

 Lifestyle2 months ago
Lifestyle2 months agoJump for Good: San Clemente Pier Fundraiser Allows Legal Leaps
-
 Science2 months ago
Science2 months agoMars Observed: Detailed Imaging Reveals Dust Avalanche Dynamics
-

 Health2 months ago
Health2 months agoResearcher Uncovers Zika Virus Pathway to Placenta Using Nanotubes
-

 Entertainment2 months ago
Entertainment2 months agoJennifer Lopez Addresses A-Rod Split in Candid Interview
-

 World2 months ago
World2 months agoUS Passport Ranks Drop Out of Top 10 for First Time Ever