Science
Breakthrough in RNA Folding Simulation Enhances Therapeutic Research
Researchers at Tokyo University of Science have made significant advances in understanding RNA folding through molecular dynamics simulations, according to a study published on November 4, 2025, in the journal ACS Omega. This study addresses the complexities of RNA folding, which is crucial for developing RNA-based therapeutics.
RNA is not just a messenger for genetic information; it plays vital roles in gene regulation and processing across all living systems. Its ability to adopt complex three-dimensional shapes is essential for its diverse functions. As the demand for RNA-based therapies grows, accurately predicting these shapes has become increasingly important for biotechnology.
Advancements in Simulation Techniques
Despite the importance of RNA folding, accurately simulating the folding process from an unfolded state has posed a challenge in computational biophysics. Traditional molecular dynamics (MD) simulations have struggled to achieve consistent results, particularly for complex RNA structures. Historically, most studies have only successfully modeled simple stem-loop structures.
In response to this challenge, Associate Professor Tadashi Ando from the Department of Applied Electronics at the Tokyo University of Science conducted a comprehensive evaluation of modern simulation methods. His research aimed to combine advanced computational tools to model a significantly larger and structurally diverse library of RNA stem loops.
The study incorporated two cutting-edge components: the DESRES-RNA atomistic force field, designed for high precision in RNA simulations, and the GB-neck2 implicit solvent model. The implicit solvent model approximates liquid environments as a continuous medium, which accelerates conformational sampling compared to traditional explicit solvent models.
Successful Folding of RNA Structures
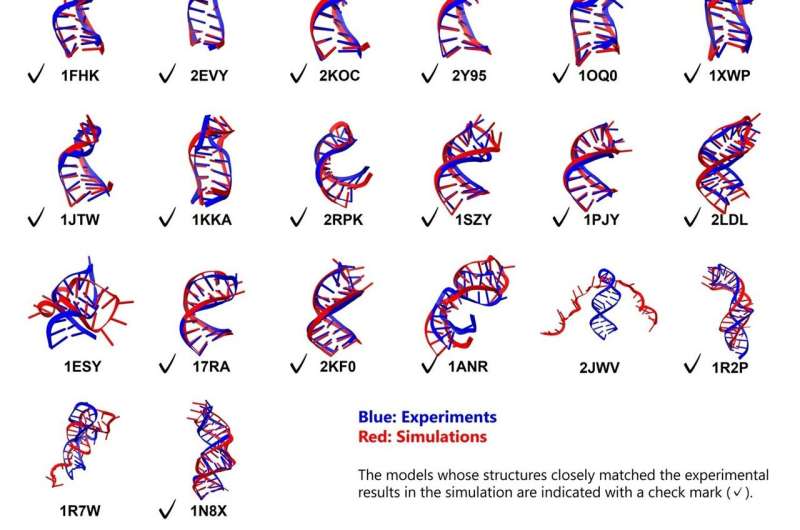
Dr. Ando applied his methodology to a diverse set of 26 RNA stem loops, ranging in length from 10 to 36 nucleotides and featuring various structural elements like bulges and internal loops. All simulations commenced from fully extended, unfolded conformations. The results exhibited a remarkable degree of folding stability and accuracy, with 23 out of 26 RNA molecules achieving their expected structures.
For the 18 simpler structures, folding accuracy was exceptional, with root mean square deviation (RMSD) values—an indicator of deviation from known experimental structures—of less than 2 Å for the stem regions and less than 5 Å overall. Among the more complex motifs, five out of eight containing bulges or internal loops were successfully folded, revealing distinct folding pathways.
“Previous studies have reported only two or three folding examples of simple stem-loop structures of approximately 10 residues, whereas this study deals with structures of much greater scale and complexity,”
Dr. Ando stated, emphasizing the significance of these findings in the field of computational biology.
The successful simulation of a broad and complex library of RNA structures marks a crucial milestone, validating the chosen combination of force field and solvent models. This research not only enhances the understanding of RNA folding but also sets the stage for future refinements in simulation tools.
While the study achieved high accuracy in stem regions, the loop regions demonstrated less precision, with RMSD values around 4 Å. This suggests a need for further optimization of the implicit solvent model parameters, particularly in modeling non-canonical base pairs and the influence of divalent cations like magnesium.
Understanding RNA folding is essential for designing drugs that can target RNA effectively, which could lead to innovative treatments for genetic disorders, viral infections such as COVID-19 and influenza, as well as certain cancers. “The ability to reproduce the overall folding of the basic stem-loop structure is an important milestone in understanding and predicting the structure, dynamics, and function of RNA using highly accurate and reliable computational models,” Dr. Ando concluded. He anticipates that these computational techniques will significantly impact RNA molecule design and drug discovery.
The implications of this research are profound, potentially paving the way for new medical tools and therapeutic strategies in the future.
-
 Science3 weeks ago
Science3 weeks agoInterstellar Object 3I/ATLAS Emits Unique Metal Alloy, Says Scientist
-

 Politics3 weeks ago
Politics3 weeks agoAfghan Refugee Detained by ICE After Asylum Hearing in New York
-

 Business3 weeks ago
Business3 weeks agoIconic Sand Dollar Social Club Listed for $3 Million in Folly Beach
-

 Health3 weeks ago
Health3 weeks agoPeptilogics Secures $78 Million to Combat Prosthetic Joint Infections
-

 Science3 weeks ago
Science3 weeks agoResearchers Achieve Fastest Genome Sequencing in Under Four Hours
-

 Lifestyle3 weeks ago
Lifestyle3 weeks agoJump for Good: San Clemente Pier Fundraiser Allows Legal Leaps
-

 Health3 weeks ago
Health3 weeks agoResearcher Uncovers Zika Virus Pathway to Placenta Using Nanotubes
-
 Science3 weeks ago
Science3 weeks agoMars Observed: Detailed Imaging Reveals Dust Avalanche Dynamics
-

 World3 weeks ago
World3 weeks agoUS Passport Ranks Drop Out of Top 10 for First Time Ever
-

 Entertainment3 weeks ago
Entertainment3 weeks agoJennifer Lopez Addresses A-Rod Split in Candid Interview
-

 Business3 weeks ago
Business3 weeks agoSan Jose High-Rise Faces Foreclosure Over $182.5 Million Loan
-

 Top Stories3 weeks ago
Top Stories3 weeks agoChicago Symphony Orchestra Dazzles with Berlioz Under Mäkelä